How synthetic disease outbreaks help predict real ones
New method outperformed 75% of the models used for forecasting COVID-19

What if the spread of an infectious disease could be forecast with little real-time data, or even without any data at all? Statistical scientists at Los Alamos National Laboratory have achieved just that with a new algorithmic method called Synthetic Method of Analogues (sMOA).
The new method generates “synthetic data” prior to any disease data being observed, making it very useful at the onset of a pandemic when no historical data on an emerging pathogen are available. By first building a large library of computer-generated outbreak shapes, sMOA matches patterns that closely resemble early onset of current cases and uses the next likely scenario in these patterns to forecast the disease spread.
Read the paper
Why this matters: Forecasts from sMOA are available weeks earlier than many traditional methods that must wait for historical data to exist. Early forecasts help public health decision-makers respond faster to disease outbreaks and provide adequate staffing, vaccine inventory and operations.
What they did:
- Scientists used compartmental models to simulate many possible outbreak curves and create a synthetic library of possible scenarios.
- They took segments of early outbreak data and matched them to synthetic data that most resembled the recent outbreak behavior. By looking at what happened after the matching point in the synthetic data, they then aggregated future scenarios to obtain a forecast for a certain number of steps ahead.
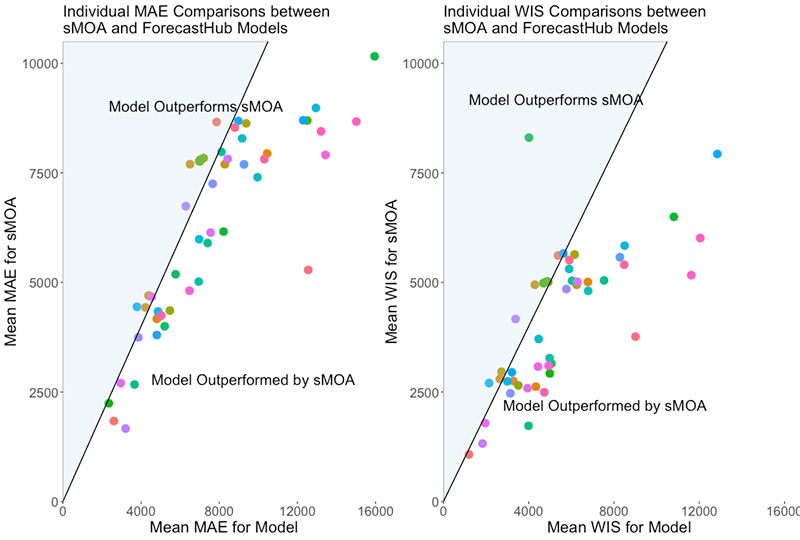
- In a retroactive study, sMOA was able to provide forecasts well before forecasts started being submitted to the COVID-19 Forecast Hub. Furthermore, sMOA outperformed most of the models even toward the end of the pandemic — all without training to historical data.
Funding: Los Alamos Laboratory Directed Research and Development, National Institutes of Health, National Institute of General Medical Sciences
LA-UR-25-30295