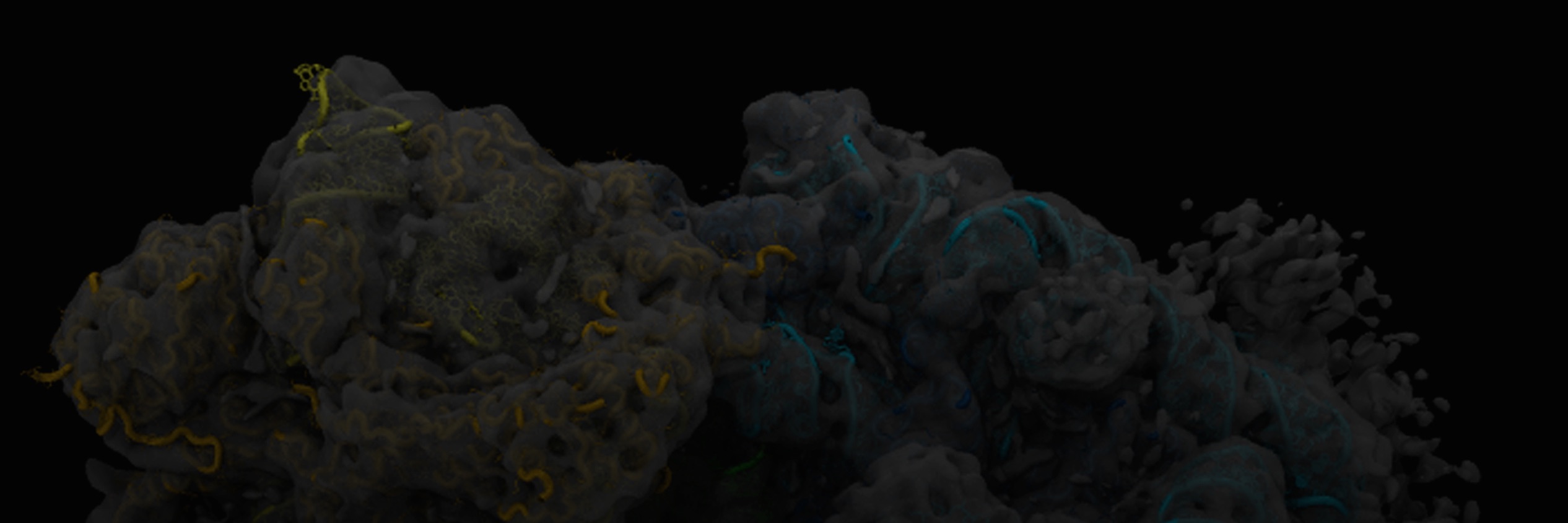
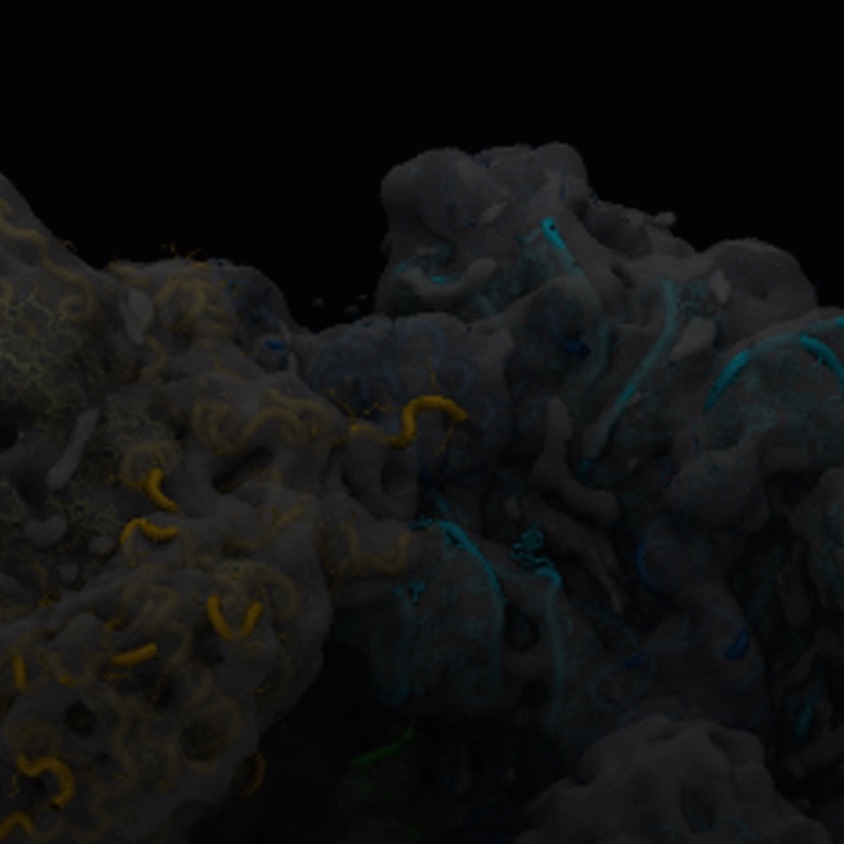
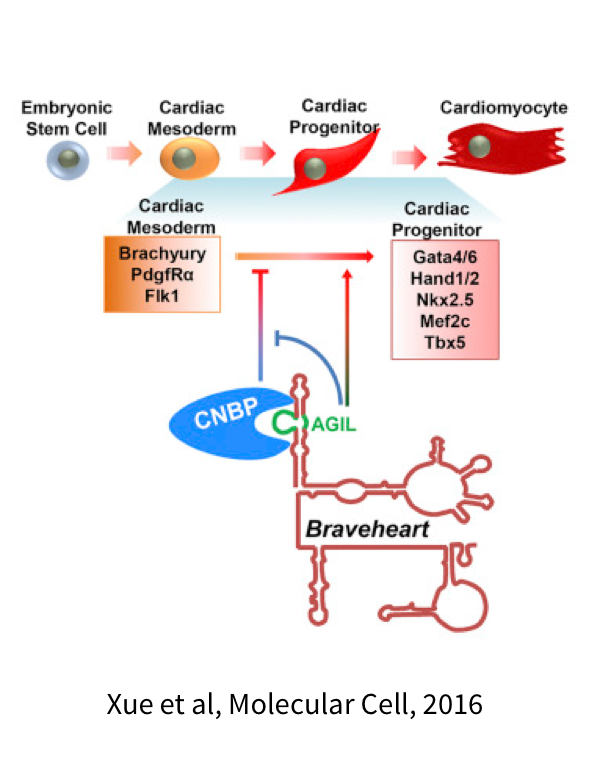
The Sanbonmatsu team uses computational and experimental approaches to understand the mechanism of a diverse array of non-coding RNA systems, including ribosomes, riboswitches and long non-coding RNAs. Originally focusing on large-scale simulations of the ribosome, we have expanded into joint computational/experimental studies of riboswitches and purely experimental studies of long non-coding RNAs. The ribosome is one of the few large RNA systems that has been studied mechanistically. We are applying our knowledge gained from ribosome studies to long non-coding RNA systems.