Biological Applications
Contact
- Harshini Mukundan
Developing technologies to detect a range of real-world diseases
Applications range from breast cancer biomarker identification, tuberculosis identification, and more recently early identification of traumatic brain injury.
Key focus areas
We have adapted the biosensor for the rapid (15 min to result), sensitive (< 200 femtomolar) and specific (low non-specific binding) detection of carcinoembryonic antigen (CEA), a cancer biomarker. Whereas the presence of this antigen in serum is indicative of several cancerous and non-cancerous conditions, its presence in the aspirate fluid extracted from the breast is a definitive indicator of breast cancer (BC). However, detection of the antigen in the protein rich nipple aspirate fluid (NAF) is a challenge because of the small sample size (1-50 microliter). We have completed a pilot study of 15 serum samples and 5 NAF samples from individuals with abnormal mammograms. The CEA concentrations measured have been sent to our collaborators at the City of Hope Hospitals in California for corroboration.
In addition, we are currently developing a multi-channel waveguide, which will allow us parallel evaluation of three different biomarkers. We hope to develop assays for a suite of BC biomarkers for simultaneous detection on the multi-channel waveguide. This will provide a more comprehensive diagnostic tool and can serve as a supplement to the current BC detection methods such as mammograms
Strain Specific Detection of Influenza Viruses using Novel Synthetic Carbohydrate Ligands
Influenza is caused by a highly pathogenic virus of the Orthomyxoviridae family. The disease accounts for over 150 million infections and 200,000 hospitalizations annually in the United States alone. The segmented genome of the virus is responsible for an unusually high mutation frequency that allows it to evade the host immune response. Early and accurate diagnosis of the specific variant of the virus would undeniably facilitate a more targeted response by health care providers. However, current rapid (~15 min) diagnostic tests for influenza lack in specificity and sensitivity. Another major issue is the use of antibodies and other reagents that are sensitive, unstable, expensive and suffer from cross-reactivity issues. The recent outbreaks of the potentially deadly H5N1 strain of the virus that have occurred in underdeveloped areas of the world highlights this problem. There is a clear need for stable synthetic recognition elements that can specifically recognize influenza virus strains.
The biosensor team at the Los Alamos National Laboratory, in collaboration with researchers at the University of Cincinnati, has developed synthetic carbohydrate ligands that can specifically recognize influenza viridae variants. Our modular synthesis strategy is based on the fact that all variants of the virus bind terminal sialic acid residues using two surface glycoproteins, namely hemagglutinin (HA) and neuraminidase (NA). We have synthesized S-sialosides resistant towards the innate enzymatic activity of NA and used them to both capture and report intact viral particles in a sandwich immunoassay format. The position of the sialic acid, its orientation, density, tether length, ancillary groups and other structure parameters can all influence binding efficiency. All these variables were easily accommodated in our synthetic strategy to generate two glycoconjugates (BG-1 and BG-2) that differ in the presentation of the sialic acid. In BG-1, the recognition element directly attached to the spacer was designed to exhibit no discrimination between viral variants, while the 2.6-linked sialic acid onto a lactose scaffold in BG-2 was expected to show differential binding.
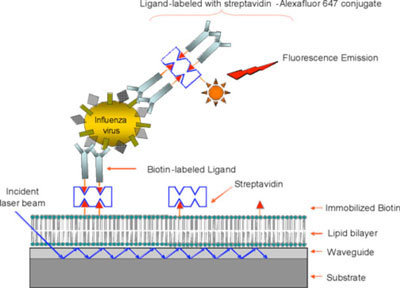
A sandwich immunoassay (where the virus is sandwiched between a biotinylated capture moiety and a fluorescently labeled reporter) was developed using these ligands on the surface of the waveguide-based optical biosensor (also developed at LANL). A schematic of the assay strategy is shown in Fig. 1. The biosensor offers a sensitive and specific assay platform for the detection of biological samples in complex biological milieu. We have previously demonstrated the efficient detection of Bacillus anthracis spores and breast cancer biomarkers using this platform. Using this assay format, we demonstrate, for the first time, a capture and reporter pair entirely based on carbohydrates, thus potentially eliminating the need for antibodies.
BG-1 can bind both H1N1 and H3N2 variants of the influenza virus confirming our expectations that this presentation of the sialic acid is not strain-specific. We then tested the binding of the H1N1 and H3N2 variants to BG-2. While A/Sydney/26/95 (H3N2) bound well to BG-2, we observed no detectable signal with A/Beijing/262/95 (H1N1) demonstrating an absolute discrimination among these strains (Fig 2). Thus, this study demonstrates the application of synthetic carbohydrate ligands to the specific detection of influenza variants for the first time.
Thus, we have developed stable S-sialoside glycoconjugates that can be used both as capture and reporter recognition elements for rapid, strain specific detection of Influenza virus. These non-natural sialic acid derivatives are especially attractive as they allow the capture of the full viral particle and are not hydrolyzed by the innate enzymatic activity of NAs present in real world samples. We have also found that these glycoconjugates are highly robust over extended time periods (> six months) at ambient temperatures and, accordingly, ideal for field assays. A particular intrinsic advantage of using glycoconjugates over antibodies is their resistance towards antigenic drifts since their binding sites are highly conserved. A focused glycan microarray comprising of stable S-sialosides could yield a fingerprint pattern of response for influenza field isolates. From the instrumentation side, we have extended our biosensor platform to demonstrate the capture of intact viral particles. We are currently developing a multi-channel adaptation of the optical biosensor that could be applied to detect Influenza from multiple samples or alternatively, multiple pathogens simultaneously. A prototype has been developed and is being evaluated.
This effort required the contribution of a diverse team of researchers including synthetic chemists, microbiologists, engineers and biologists. Dr. Jurgen Schmidt and Dr. Ramesh Kale, with Dr. Suri Iyer and Dr. Lewallen of the University of Cincinnati, were responsible for the synthesis of the glycoconjugates. Assay development and biodetection, under Dr. Basil Swanson, was performed by Dr. H. Mukundan. Dr. Jennifer Foster-Harris and Dominique Price, in Dr. Hong Cai’s laboratory, were responsible for the culture and characterization of the viruses. Kevin Grace and Karen Grace of Los Alamos were contributing engineers whose efforts that lead to the development of the biosensor platform.
In addition to breast cancer, the team is interested in the non-invasive diagnosis of tuberculosis (TB) in patient urine. With the emergence of drug resistant variants, the World Health Organization has redefined TB as a global health crisis. Yet, early diagnosis of the disease is a challenge. Preliminary studies, performed in collaboration with the National Institutes of Health, have indicated that at least three different bacterial biomarkers specific to Mycobacterium tuberculosis are secreted in the urine of patients with the multi-drug resistant TB (MDR-TB). We are currently optimizing immunoassays for these markers for application to the waveguide platform. We hope to explore the presence of biomarkers in patient urine as well as evaluate any differential expression of these biomolecules in MDR versus. regular TB. The project will definitely contribute to our understanding of TB biomarkers and may serve as a diagnostic tool for the disease.